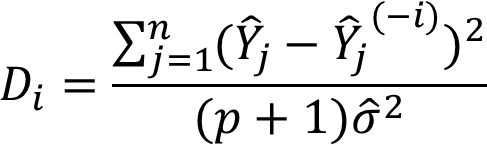DSC 3091- Advanced Statistics Applications I
Multiple Linear Regression
Dr Jagath Senarathne
Department of Statistics and Computer Science
Multiple Linear Regression
Studying the variation of Y(response variable) as a function of more than one X(explanatory) variables.
The general multiple linear regression model;
\[ Y= \beta_0+\beta_1x_1+\beta_2x_2...+\beta_px_p+ \epsilon\]
Where, \(\epsilon \sim N(0,\sigma^2)\) is the random noise component of the model and \(\beta_0,\beta_1,...,\beta_p\), are the unknown parameters.
Example 1
You might want to predict how well a stock will do based on some other information that you just happen to have.
Example 2
Imagine that you are a tourist guide. You need to provide the price range of food to your clients. The price of those food usually correlates with the Food Quality and Service Quality of the Restaurant.
Example 3
Imagine that you’re a traffic planner in your city and need to estimate the average commute time of drivers going from the east side of the city to the west. You know that it will depend on a number of factors like the distance driven, the number of stoplights on the route, and the number of other vehicles on the road.
How to fit a multiple linear regression model?
Steps you can follow:
- Graphical Interpretation
- Parameter Estimation
- Tests on Parameters
- Analysis of Variance
- Interpretation and Prediction
Graphical Interpretation
Before fitting a multiple liner regression model, plot a scatterplot between every possible pair of variables.
It helps in visualizing the strength of relationships.
Can also be used for variable selection and identifying co-linearity.
Example
Let’s consider the built-in dataset, “abalone”, in the package called “AppliedPredictiveModeling”. Abalone data explains various physical measurements of sea snails. The main objective of this study is to predict the number of rings(which is an indicator of the age of the snail) using the other variables.
Example Cont…
Draw a correlogram using corrplot() function
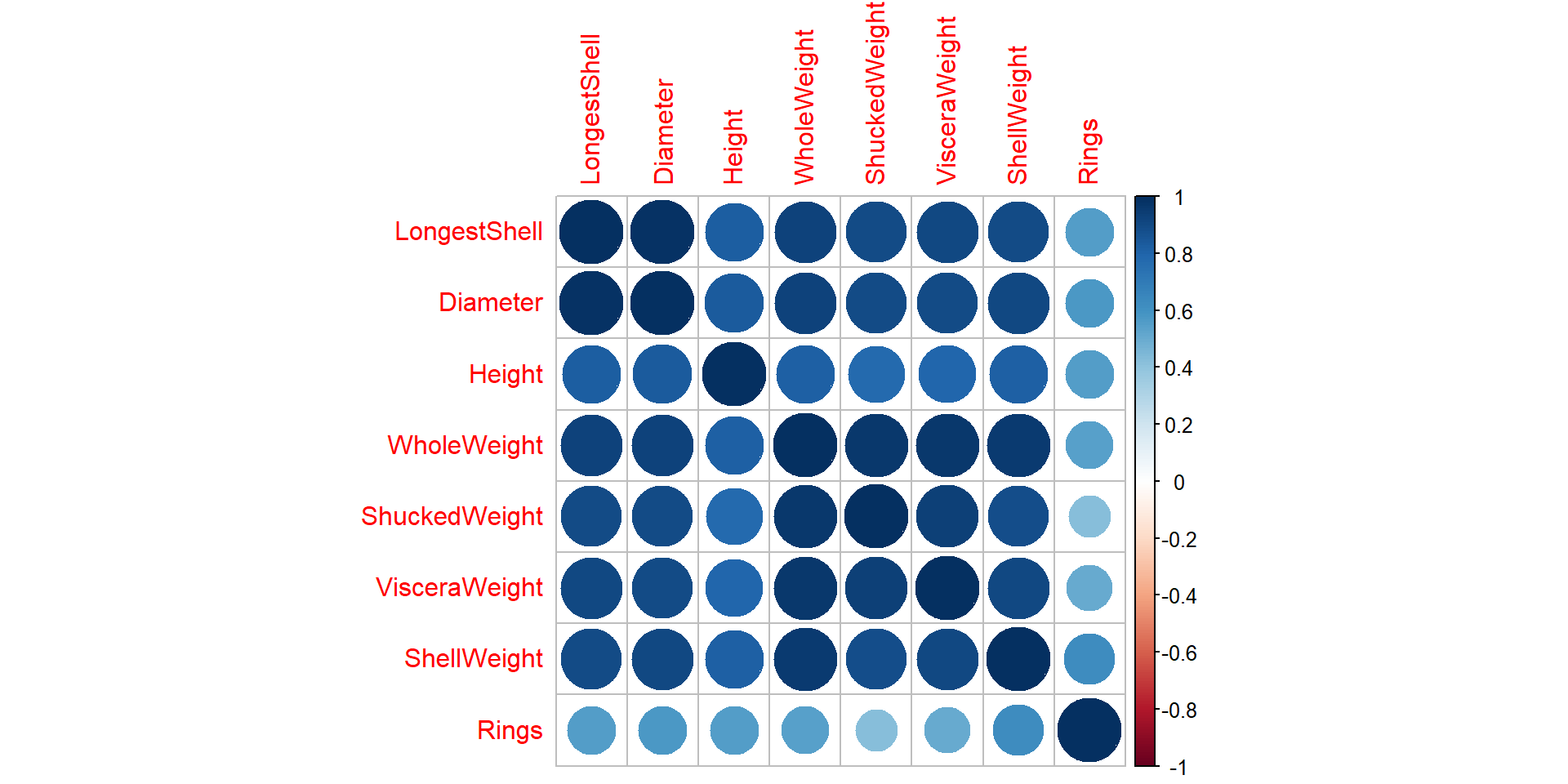
Draw scatter plots using chart.Correlation():

Parameter Estimation
- Similar to the simple linear regression, we can use
lm()function to estimate the unknown parameters \(\beta_0, \beta_1,.,\beta_p\).
model1 <- lm(Rings~as.factor(Type) +LongestShell+ Diameter+ WholeWeight+ ShuckedWeight+ VisceraWeight+ ShellWeight)
model1
Call:
lm(formula = Rings ~ as.factor(Type) + LongestShell + Diameter +
WholeWeight + ShuckedWeight + VisceraWeight + ShellWeight)
Coefficients:
(Intercept) as.factor(Type)I as.factor(Type)M LongestShell
4.24786 -0.87919 0.04437 -0.05352
Diameter WholeWeight ShuckedWeight VisceraWeight
12.73487 9.06362 -19.95631 -10.18938
ShellWeight
9.57238
Call:
lm(formula = Rings ~ as.factor(Type) + LongestShell + Diameter +
WholeWeight + ShuckedWeight + VisceraWeight + ShellWeight)
Residuals:
Min 1Q Median 3Q Max
-8.1813 -1.3130 -0.3437 0.8690 13.7654
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 4.24786 0.28883 14.707 < 2e-16 ***
as.factor(Type)I -0.87919 0.10269 -8.562 < 2e-16 ***
as.factor(Type)M 0.04437 0.08380 0.529 0.597
LongestShell -0.05352 1.81860 -0.029 0.977
Diameter 12.73487 2.22738 5.717 1.16e-08 ***
WholeWeight 9.06362 0.72947 12.425 < 2e-16 ***
ShuckedWeight -19.95631 0.82169 -24.287 < 2e-16 ***
VisceraWeight -10.18938 1.29997 -7.838 5.76e-15 ***
ShellWeight 9.57238 1.12490 8.510 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 2.207 on 4168 degrees of freedom
Multiple R-squared: 0.5324, Adjusted R-squared: 0.5315
F-statistic: 593.3 on 8 and 4168 DF, p-value: < 2.2e-16Let’s create a dummy variable!
abalone$Type.I=ifelse(abalone$Type=="I",1,0)
abalone$Type.M=ifelse(abalone$Type=="M",1,0)
model2 <- lm(Rings~Type.I + Type.M+ LongestShell+ Diameter+ WholeWeight+ ShuckedWeight+ VisceraWeight+ ShellWeight,data=abalone)
summary(model2)
Call:
lm(formula = Rings ~ Type.I + Type.M + LongestShell + Diameter +
WholeWeight + ShuckedWeight + VisceraWeight + ShellWeight,
data = abalone)
Residuals:
Min 1Q Median 3Q Max
-8.1813 -1.3130 -0.3437 0.8690 13.7654
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 4.24786 0.28883 14.707 < 2e-16 ***
Type.I -0.87919 0.10269 -8.562 < 2e-16 ***
Type.M 0.04437 0.08380 0.529 0.597
LongestShell -0.05352 1.81860 -0.029 0.977
Diameter 12.73487 2.22738 5.717 1.16e-08 ***
WholeWeight 9.06362 0.72947 12.425 < 2e-16 ***
ShuckedWeight -19.95631 0.82169 -24.287 < 2e-16 ***
VisceraWeight -10.18938 1.29997 -7.838 5.76e-15 ***
ShellWeight 9.57238 1.12490 8.510 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 2.207 on 4168 degrees of freedom
Multiple R-squared: 0.5324, Adjusted R-squared: 0.5315
F-statistic: 593.3 on 8 and 4168 DF, p-value: < 2.2e-16Methods of building a multiple linear regression model
- Backward Elimination
- Forward Selection
- Bidirectional Elimination (Stepwise regression)
Backward elimination
Set a significance level for which data will stay in the model.
Next, fit the full model with all possible predictors.
Consider the predictor with the highest P-value. If the P-value is greater than the significance level, you’ll move to step four, otherwise, you’re done!
Remove that predictor with the highest P-value.
Fit the model without that predictor variable.
Go back to step 3, do it all over, and keep doing that until you come to a point where even the highest P-value is < SL.
model2 <- lm(Rings~Type.I + Type.M+ LongestShell+ Diameter+ WholeWeight+ ShuckedWeight+ VisceraWeight+ ShellWeight,data=abalone)
step.model <- step(model2, direction = "backward",trace=FALSE)
summary(step.model)
Call:
lm(formula = Rings ~ Type.I + Diameter + WholeWeight + ShuckedWeight +
VisceraWeight + ShellWeight, data = abalone)
Residuals:
Min 1Q Median 3Q Max
-8.1597 -1.3126 -0.3417 0.8697 13.7453
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 4.27930 0.26379 16.222 < 2e-16 ***
Type.I -0.90566 0.08966 -10.101 < 2e-16 ***
Diameter 12.64787 0.94590 13.371 < 2e-16 ***
WholeWeight 9.06125 0.72926 12.425 < 2e-16 ***
ShuckedWeight -19.93199 0.81772 -24.375 < 2e-16 ***
VisceraWeight -10.22104 1.29262 -7.907 3.34e-15 ***
ShellWeight 9.57105 1.12421 8.514 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 2.206 on 4170 degrees of freedom
Multiple R-squared: 0.5324, Adjusted R-squared: 0.5317
F-statistic: 791.3 on 6 and 4170 DF, p-value: < 2.2e-16Backward elimination using SignifReg package
library(SignifReg)
nullmodel = lm(Rings~1, data=abalone)
fullmodel = lm(Rings~Type.I + Type.M+ LongestShell+ Diameter+ WholeWeight+ ShuckedWeight+ VisceraWeight+ ShellWeight, data=abalone)
scope = list(lower=formula(nullmodel),upper=formula(fullmodel))
Sub.model=drop1SignifReg(fullmodel, scope, alpha = 0.05, criterion = "p-value",override="TRUE")
summary(Sub.model)
Call:
lm(formula = Rings ~ Type.I + Type.M + LongestShell + Diameter +
WholeWeight + VisceraWeight + ShellWeight, data = abalone)
Residuals:
Min 1Q Median 3Q Max
-8.7218 -1.4383 -0.4426 0.8413 16.3379
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 5.19685 0.30571 16.999 < 2e-16 ***
Type.I -1.08469 0.10933 -9.921 < 2e-16 ***
Type.M -0.07688 0.08937 -0.860 0.3897
LongestShell -3.36753 1.93732 -1.738 0.0822 .
Diameter 13.40996 2.37930 5.636 1.85e-08 ***
WholeWeight -5.61381 0.43643 -12.863 < 2e-16 ***
VisceraWeight -1.87624 1.33974 -1.400 0.1615
ShellWeight 26.79546 0.93284 28.725 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 2.357 on 4169 degrees of freedom
Multiple R-squared: 0.4663, Adjusted R-squared: 0.4654
F-statistic: 520.3 on 7 and 4169 DF, p-value: < 2.2e-16Forward selection
- Choose the significance level (SL = 0.05).
- Fit all possible simple regression models and select the one with the lowest P-value.
- Keep this variable and fit all possible models with one extra predictor added to the one you already have.
- Find the predictor with the lowest P-value. If P < Sl, go back to step 3. Otherwise, you’re done!
model2 <- lm(Rings~Type.I + Type.M+ LongestShell+ Diameter+ WholeWeight+ ShuckedWeight+ VisceraWeight+ ShellWeight,data=abalone)
step.model2 <- step(model2, direction = "forward",trace=FALSE)
summary(step.model2)
Call:
lm(formula = Rings ~ Type.I + Type.M + LongestShell + Diameter +
WholeWeight + ShuckedWeight + VisceraWeight + ShellWeight,
data = abalone)
Residuals:
Min 1Q Median 3Q Max
-8.1813 -1.3130 -0.3437 0.8690 13.7654
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 4.24786 0.28883 14.707 < 2e-16 ***
Type.I -0.87919 0.10269 -8.562 < 2e-16 ***
Type.M 0.04437 0.08380 0.529 0.597
LongestShell -0.05352 1.81860 -0.029 0.977
Diameter 12.73487 2.22738 5.717 1.16e-08 ***
WholeWeight 9.06362 0.72947 12.425 < 2e-16 ***
ShuckedWeight -19.95631 0.82169 -24.287 < 2e-16 ***
VisceraWeight -10.18938 1.29997 -7.838 5.76e-15 ***
ShellWeight 9.57238 1.12490 8.510 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 2.207 on 4168 degrees of freedom
Multiple R-squared: 0.5324, Adjusted R-squared: 0.5315
F-statistic: 593.3 on 8 and 4168 DF, p-value: < 2.2e-16Forward selection using SignifReg package
library(SignifReg)
nullmodel = lm(Rings~1, data=abalone)
fullmodel = lm(Rings~Type.I + Type.M+ LongestShell+ Diameter+ WholeWeight+ ShuckedWeight+ VisceraWeight+ ShellWeight, data=abalone)
scope = list(lower=formula(nullmodel),upper=formula(fullmodel))
sub.model2=add1SignifReg(nullmodel,scope, alpha = 0.05, criterion = "p-value",override="TRUE")
summary(sub.model2)
Call:
lm(formula = Rings ~ LongestShell, data = abalone)
Residuals:
Min 1Q Median 3Q Max
-5.9665 -1.6961 -0.7423 0.8733 16.6776
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 2.1019 0.1855 11.33 <2e-16 ***
LongestShell 14.9464 0.3452 43.30 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 2.679 on 4175 degrees of freedom
Multiple R-squared: 0.3099, Adjusted R-squared: 0.3098
F-statistic: 1875 on 1 and 4175 DF, p-value: < 2.2e-16Stepwise regression
Select a significance level to enter and a significance level to stay.
Perform the next step of forward selection where you add the new variable.
Now perform all of the steps of backward elimination.
Now head back to step two, then move forward to step 3, and so on until no new variables can enter and no new variables can exit.
model2 <- lm(Rings~Type.I + Type.M+ LongestShell+ Diameter+ WholeWeight+ ShuckedWeight+ VisceraWeight+ ShellWeight,data=abalone)
step.model2 <- step(model2, direction = "forward", trace=FALSE)
summary(step.model2)
Call:
lm(formula = Rings ~ Type.I + Type.M + LongestShell + Diameter +
WholeWeight + ShuckedWeight + VisceraWeight + ShellWeight,
data = abalone)
Residuals:
Min 1Q Median 3Q Max
-8.1813 -1.3130 -0.3437 0.8690 13.7654
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 4.24786 0.28883 14.707 < 2e-16 ***
Type.I -0.87919 0.10269 -8.562 < 2e-16 ***
Type.M 0.04437 0.08380 0.529 0.597
LongestShell -0.05352 1.81860 -0.029 0.977
Diameter 12.73487 2.22738 5.717 1.16e-08 ***
WholeWeight 9.06362 0.72947 12.425 < 2e-16 ***
ShuckedWeight -19.95631 0.82169 -24.287 < 2e-16 ***
VisceraWeight -10.18938 1.29997 -7.838 5.76e-15 ***
ShellWeight 9.57238 1.12490 8.510 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 2.207 on 4168 degrees of freedom
Multiple R-squared: 0.5324, Adjusted R-squared: 0.5315
F-statistic: 593.3 on 8 and 4168 DF, p-value: < 2.2e-16Model Diagnosis
The model fitting is just the first part of the regression analysis since this is all based on certain assumptions.
Regression diagnostics are used to evaluate the model assumptions and investigate whether or not there are observations with a large, undue influence on the analysis.
Model Assumptions
Before using a regression model for interpreting the data, we must check that the model assumptions are met.
Basic assumptions of regression models:
Linearity of the data: The relationship between the predictor (x) and the outcome (y) is assumed to be linear.
Normality of residuals: The residual errors are assumed to be normally distributed.
Homogeneity of residuals variance: The residuals are assumed to have a constant variance (homoscedasticity)
Independence of residuals error terms.
Checking Assumptions
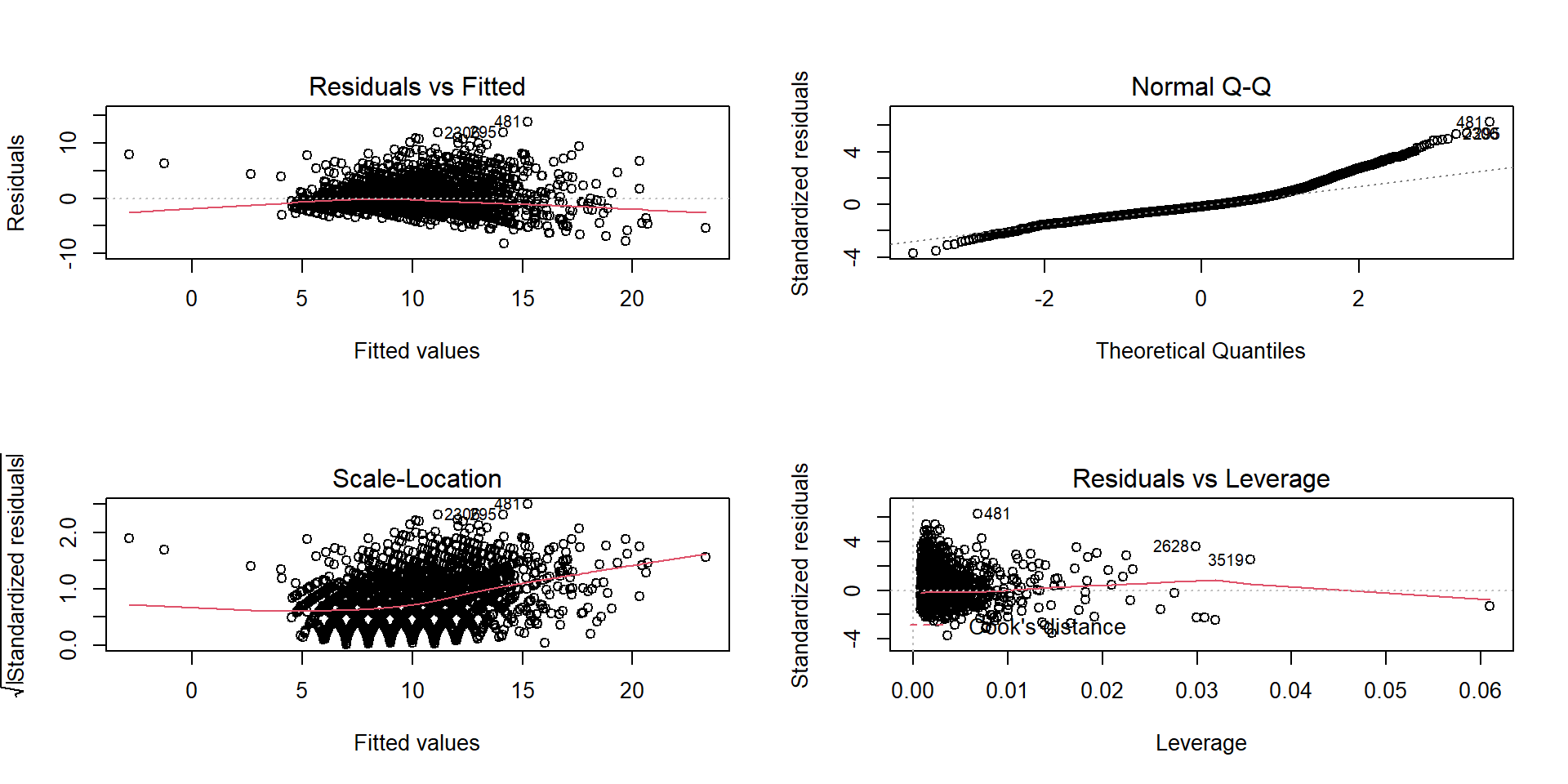
Using ggfortify package
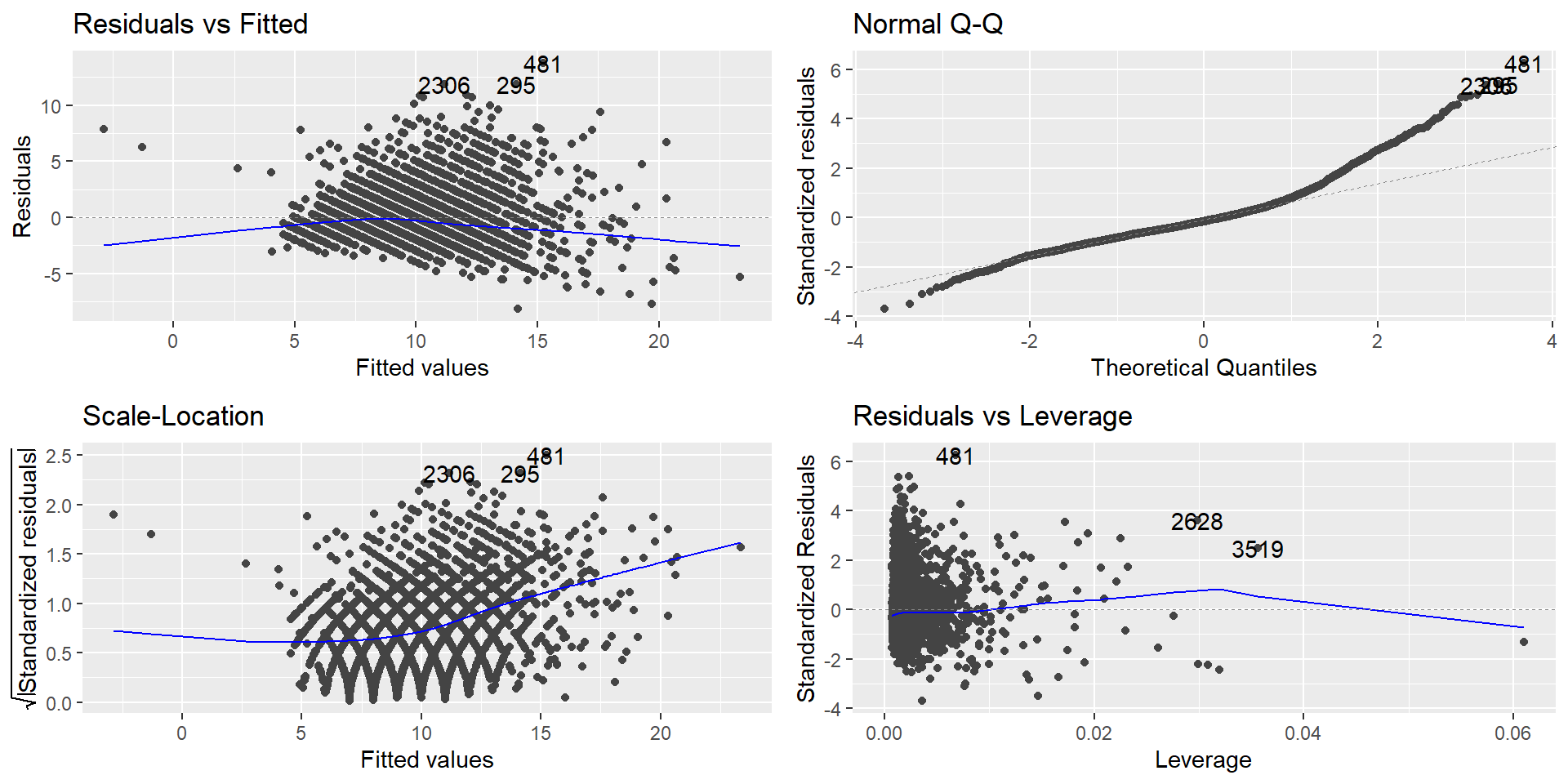
The diagnostic plots show residuals in four different ways:
Residuals vs Fitted: to check the linear relationship assumptions.
Normal Q-Q: to examine whether the residuals are normally distributed.
Scale-Location (or Spread-Location): to check the homogeneity of variance of the residuals (homoscedasticity).
Residuals vs Leverage: to identify extreme values that might influence the regression analysis.
Identifying Influential Outliers
An outlier is a point which is often distant from the others.
It is a point with a large value of its residual.
Different residual measures can be used to identify outliers.
Standardised residuals
Studentized residuals
Cook’s distance
Standardised residuals
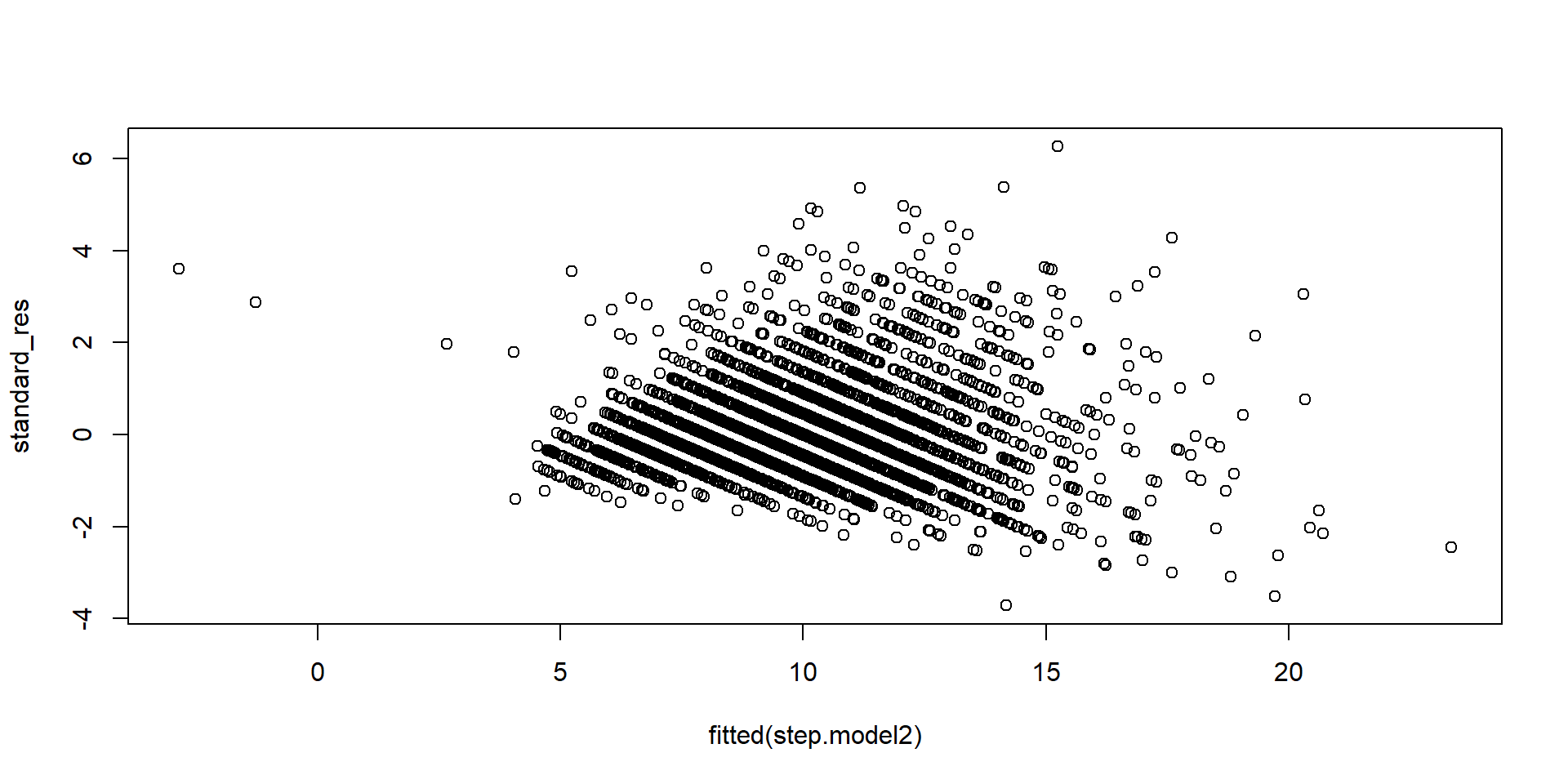
- Standardized residuals are “mostly” between 2 and 2, but they are dependent.
Studentized residuals

- For an outlier the absolute values of its Studentized residual is greater than the 0.975th quantile of the Student distribution with \(n-p-2\) degrees of freedom
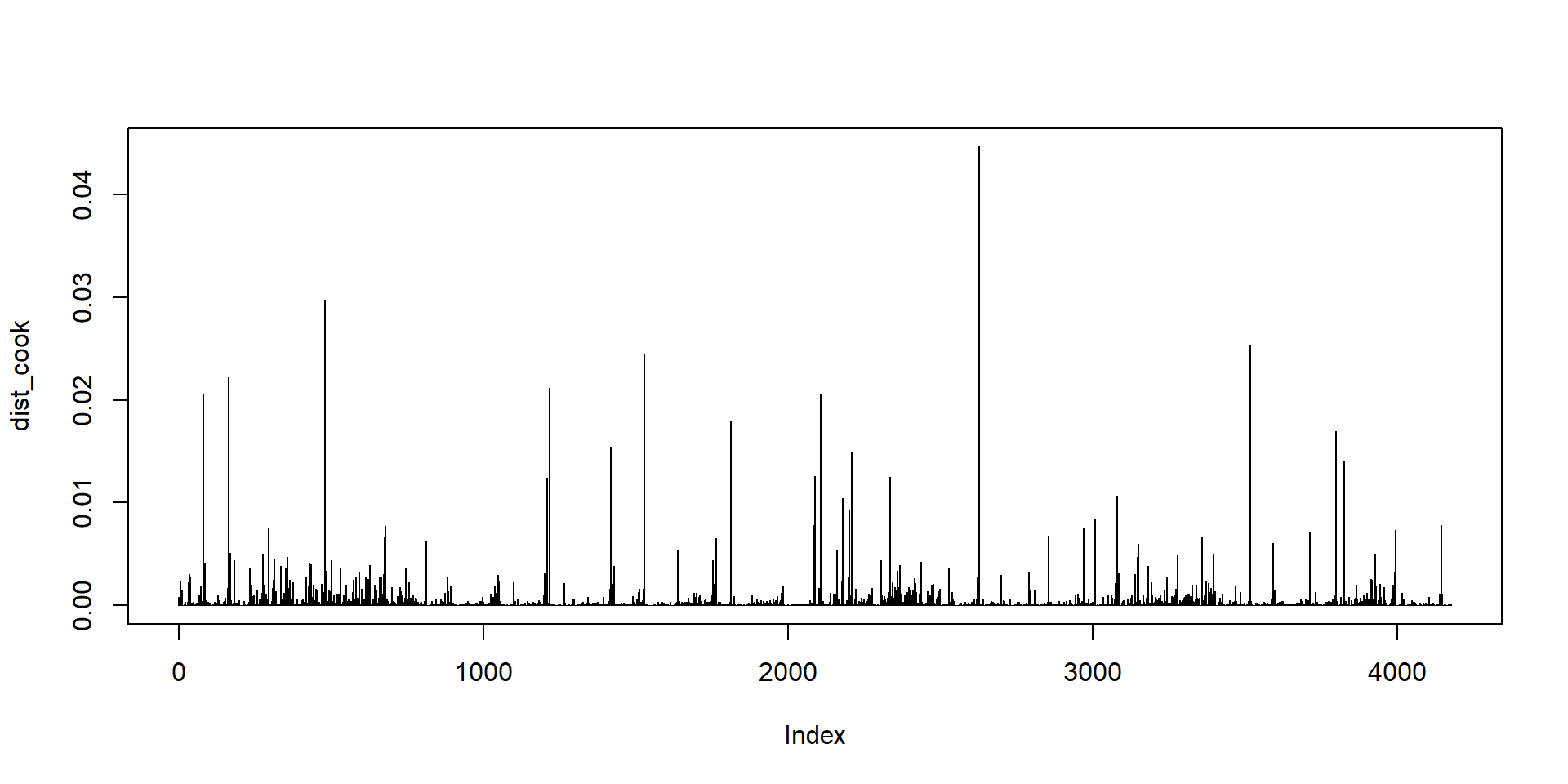
Cook’s Distance
It is calculated by removing the \(i\)th data point from the model and refitting the regression model.
It summarizes how much all the values in the regression model change when the \(i\)th observation is removed.
The formula for Cook’s distance is:
![]()
An R function to calculate Cook’s distance is
cooks.distance().
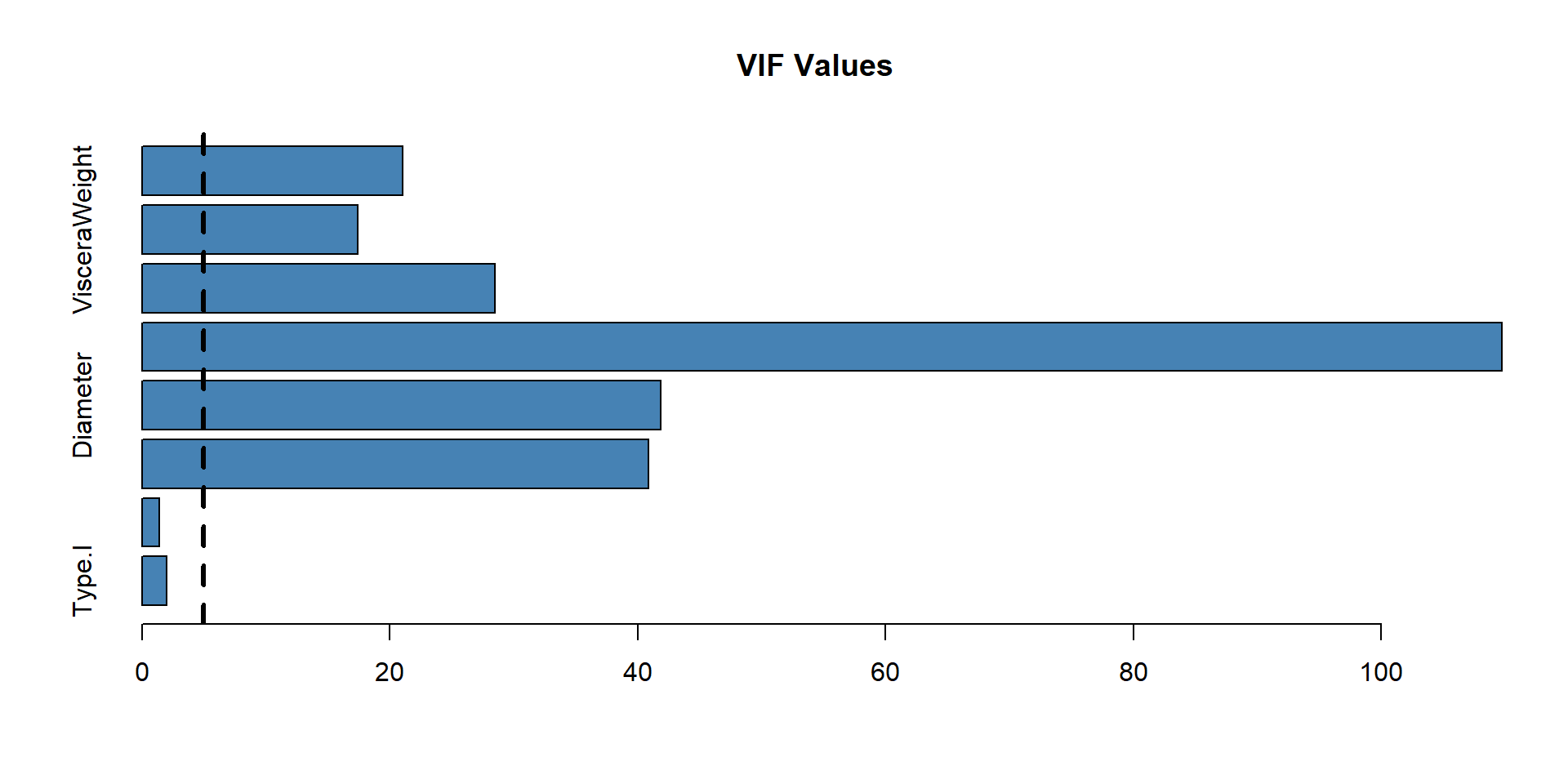
Checking for multicollinearity
It is a statistical terminology where more than one independent variable is correlated with each other.
Results in reducing the reliability of statistical inferences.
Hence, these variables must be removed when building a multiple regression model.
Variance inflation factor (VIF) is used for detecting the multicollinearity in a model.
Variance inflation factor
Testing the multicollinearity among the predictor variables
- The
mctestpackage in R provides the Farrar-Glauber test and other relevant tests for multicollinearity.
Call:
omcdiag(mod = step.model2)
Overall Multicollinearity Diagnostics
MC Results detection
Determinant |X'X|: 0.0000 1
Farrar Chi-Square: 62488.4404 1
Red Indicator: 0.7378 1
Sum of Lambda Inverse: 262.8746 1
Theil's Method: 2.8529 1
Condition Number: 104.1100 1
1 --> COLLINEARITY is detected by the test
0 --> COLLINEARITY is not detected by the test
Call:
imcdiag(mod = step.model2)
All Individual Multicollinearity Diagnostics Result
VIF TOL Wi Fi Leamer CVIF Klein
Type.I 1.9723 0.5070 579.0942 675.7720 0.7120 -0.9408 0
Type.M 1.3975 0.7155 236.7649 276.2919 0.8459 -0.6666 0
LongestShell 40.9040 0.0244 23765.6736 27733.2698 0.1564 -19.5107 1
Diameter 41.9003 0.0239 24359.0324 28425.6879 0.1545 -19.9859 1
WholeWeight 109.7357 0.0091 64759.8525 75571.2838 0.0955 -52.3425 1
ShuckedWeight 28.5255 0.0351 16393.4264 19130.2517 0.1872 -13.6063 1
VisceraWeight 17.4123 0.0574 9774.6981 11406.5499 0.2396 -8.3054 1
ShellWeight 21.0270 0.0476 11927.5022 13918.7571 0.2181 -10.0296 1
IND1 IND2
Type.I 0.0009 0.5994
Type.M 0.0012 0.3458
LongestShell 0.0000 1.1861
Diameter 0.0000 1.1868
WholeWeight 0.0000 1.2047
ShuckedWeight 0.0001 1.1732
VisceraWeight 0.0001 1.1460
ShellWeight 0.0001 1.1580
1 --> COLLINEARITY is detected by the test
0 --> COLLINEARITY is not detected by the test
Type.M , LongestShell , coefficient(s) are non-significant may be due to multicollinearity
R-square of y on all x: 0.5324
* use method argument to check which regressors may be the reason of collinearity
===================================Model Selection
Model selection criteria refer to a set of exploratory tools for improving regression models.
Each model selection tool involves selecting a subset of possible predictor variables that still account well for the variation in the regression model’s response variable.
This suggests that we need a quality criteria that considers the size of the model, since our preference is for small models that still fit well.
Several criteria are available to measure the performance of model. (AIC, BIC , \(R^2\) (Coefficient of determination), adjacent \(R^2\))
- The
summary()function gives both \(R^2\) and adjacent \(R^2\) measures together.
Call:
lm(formula = Rings ~ Type.I + Type.M + LongestShell + Diameter +
WholeWeight + ShuckedWeight + VisceraWeight + ShellWeight,
data = abalone)
Residuals:
Min 1Q Median 3Q Max
-8.1813 -1.3130 -0.3437 0.8690 13.7654
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 4.24786 0.28883 14.707 < 2e-16 ***
Type.I -0.87919 0.10269 -8.562 < 2e-16 ***
Type.M 0.04437 0.08380 0.529 0.597
LongestShell -0.05352 1.81860 -0.029 0.977
Diameter 12.73487 2.22738 5.717 1.16e-08 ***
WholeWeight 9.06362 0.72947 12.425 < 2e-16 ***
ShuckedWeight -19.95631 0.82169 -24.287 < 2e-16 ***
VisceraWeight -10.18938 1.29997 -7.838 5.76e-15 ***
ShellWeight 9.57238 1.12490 8.510 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 2.207 on 4168 degrees of freedom
Multiple R-squared: 0.5324, Adjusted R-squared: 0.5315
F-statistic: 593.3 on 8 and 4168 DF, p-value: < 2.2e-16- Choose the model with lower AIC and BIC values.
In-Class Assignment
Obtain a multiple linear regression model that best describes the variations in the response variable Rings in the abalone dataset. Briefly discuss the steps you followed and the suitability of the model you selected.